Micro-RNAs in Human Placenta: Tiny Molecules, Immense Power
Abstract
:1. Introduction
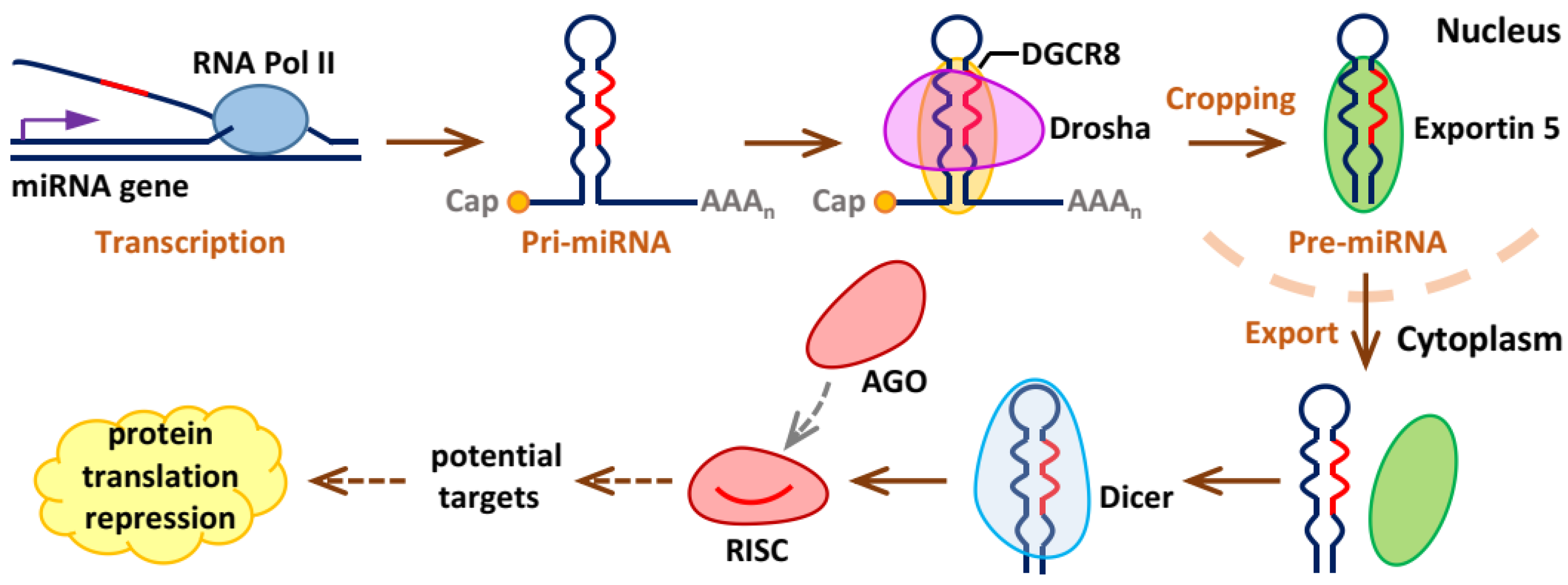
2. Biogenesis and Working Mechanisms of miRNAs
3. Human Placental Development and Structure
4. Detection of miRNAs in Human Placenta
5. Expression of Micro-RNAs in Human Placenta
| Factors | Affected miRNAs in Human Trophoblasts | ||
|---|---|---|---|
| Hypoxia | Up-regulated | miR-210 [81], miR-141 [82], miR-218 [83] | |
| Environment Factors | Phthalate | Up-regulated | miR-17-5p [88], miR-155-5p [88], miR-126-3p [88], miR-16 [89] |
| Bisphenol A | Up-regulated | miR-146a [90] | |
| Phenol | Up-regulated | miR-142-3p [91], miR15a-5p [91], miR-185 [91] | |
| Epigenetic Regulation | Methylation | Up-regulated | C19MC [95] |
| Single-Nucleotide Polymorphism (SNP) | - | 27 miRNAs [96], miR-126 [97], miR-143 [97], miR-143 [97] | |
6. Micro-RNAs and Human Gestational Disorders
6.1. Preeclampsia (PE)
| Disorder Name | MiRNAs (Reference) | ||
|---|---|---|---|
| Pre-eclampsia (PE) | Up-regulated | proliferation ↓ | miR-155 [107], miR-20a [132], miR-16 [135], miR-675 [136], miR-142-3p [138], miR-200p-3b [140], miR-137 [141], miR-146a [143] |
| migration ↓ | miR-155 [107], miR-431 [116], miR-20b [133], miR-16 [135], miR-125b [139], miR-200p-3b [140], miR-137 [141], miR-146a [143] | ||
| invasion ↓ | miR-431 [116], miR-30a-3p [117], miR-20a [132], miR-20b [133], miR-141 [137], miR-142-3p [138], miR-125b [139], miR-146a [143], miR-517a/b [145], miR-517c [145] | ||
| apoptosis ↑ | miR-30a-3p [117], miR-29b [134], miR-16 [135], miR-200p-3b [140] | ||
| differentiation ↓ | miR-17~92 clusters [146], miR-106a~363 clusters [146] | ||
| other/targets ↓ | miR-17 [68], miR-206 [129], miR-210 [131], miR-202-3p [142], miR-155 [144] | ||
| in exosomes | miR-222-3p [152], miR-486-1-5p [153], miR-486-2-5p [153], miR-155 [154], miR-136 [155], miR-494 [155], miR-495 [155] | ||
| Down-regulated | proliferation ↑ | miR-378a-5p [101], miR-376c [147], miR-335 [149], miR-126 [150] | |
| migration ↑ | miR-378a-5p [101], miR-3935 [124], miR-376c [147], miR-126 [150], | ||
| invasion ↑ | miR-378a-5p [101], miR-3935 [124] | ||
| apoptosis ↓ | miR-335 [149], miR-125a [151] | ||
| differentiation ↑ | miR-126 [150] | ||
| other/targets ↑ | miR-195 [148] | ||
| in exosomes | miR-153-3p [152], miR-653-5p [152], miR-325 [152] | ||
| Fetal Growth Restriction (FGR) | Up-regulated | oxidative stress ↓ | miR-199a-5p [161] |
| other/targets ↓ | miR-424 [162], miR-519a [163], miR-1323 [164], miR-516b [164], miR-515-5p [164], miR-520h [164], miR-519d [164], miR-526b [164] | ||
| Down-regulated | other/targets ↑ | miR-518b [163], miR-16 [165], miR-21 [165] | |
| Gestational Diabetes Mellitus (GDM) | Up-regulated | other/targets ↓ | miR-98 [166], miR-518d [167], miR-144 [168] |
| in exosomes | miR-122-5p [26], miR-132-3p [26], miR-1323 [26], miR-136-5p [26], miR-182-3p [26], miR-210-3p [26], miR-29a-3p [26], miR-29b-3p [26], miR-342-3p [26], miR-520h [26], miR-330-3p [169] | ||
| Down-regulated | proliferation ↑ | miR-296 [170], miR-96 [171], miR-137 [172] | |
| migration ↑ | miR-296 [170] | ||
| invasion ↑ | miR-296 [170] | ||
| in exosomes | miR-125b [168], miR-516-5p [173], miR-517-3p [173], miR-518-5p [173], miR-222-3p [173], miR-16-5p [173] | ||
| Early Recurrent Miscarriage | Up-regulated | apoptosis ↑ | miR-365 [174], miR-149 [175] |
| other/targets ↓ | miR-4497 [176] | ||
| Down-regulated | proliferation ↑ | miR-155-5p [177] | |
| apoptosis ↓ | miR-155-5p [177] | ||
| Preterm Birth (PTB) | Down-regulated | other/targets ↑ | miR-338 [178], miR-29b-3p [179] |
| Small-for-Gestational-Age (SGA) | Down-regulated | other/targets ↑ | miR-21 [165], miR-16 [165] |
6.2. Fetal Growth Restriction (FGR)
6.3. Gestational Diabetes Mellitus (GDM)
6.4. Other Disorders
7. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bhatti, G.K.; Khullar, N.; Sidhu, I.S.; Navik, U.S.; Reddy, A.P.; Reddy, P.H.; Bhatti, J.S. Emerging role of non-coding RNA in health and disease. Metab. Brain Dis. 2021, 36, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandao, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, B. Recent advances in the regulation of plant miRNA biogenesis. RNA Biol. 2021, 18, 2087–2096. [Google Scholar] [CrossRef]
- Do, D.N.; Dudemaine, P.L.; Mathur, M.; Suravajhala, P.; Zhao, X.; Ibeagha-Awemu, E.M. miRNA Regulatory Functions in Farm Animal Diseases, and Biomarker Potentials for Effective Therapies. Int. J. Mol. Sci. 2021, 22, 3080. [Google Scholar] [CrossRef]
- Avital, G.; Franca, G.S.; Yanai, I. Bimodal Evolutionary Developmental miRNA Program in Animal Embryogenesis. Mol. Biol. Evol. 2018, 35, 646–654. [Google Scholar] [CrossRef]
- Turco, M.Y.; Moffett, A. Development of the human placenta. Development 2019, 146, dev163428. [Google Scholar] [CrossRef]
- Hoffmann, A.; Spengler, D. The Mitochondrion as Potential Interface in Early-Life Stress Brain Programming. Front. Behav. Neurosci. 2018, 12, 306. [Google Scholar] [CrossRef]
- Tang, C.; Pan, Y.; Luo, H.; Xiong, W.; Zhu, H.; Ruan, H.; Wang, J.; Zou, C.; Tang, L.; Iguchi, T.; et al. Hedgehog signaling stimulates the conversion of cholesterol to steroids. Cell. Signal. 2015, 27, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Cui, C.; Wu, C.; Wang, J.; Zheng, X.; Ma, Z.; Zhu, P.; Guan, W.; Zhang, S.; Chen, F. Leucine supplementation during late gestation globally alters placental metabolism and nutrient transport via modulation of the PI3K/AKT/mTOR signaling pathway in sows. Food Funct. 2022, 13, 2083–2097. [Google Scholar] [CrossRef] [PubMed]
- Soncin, F.; Parast, M.M. Role of Hippo signaling pathway in early placental development. Proc. Natl. Acad. Sci. USA 2020, 117, 20354–20356. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Kim, S.C.; Park, M.N.; Jeong, J.S.; Yang, S.Y.; Lee, Y.J.; Bae, O.N.; Yang, H.S.; Seo, S.; Lee, K.S.; et al. Expression of steroidogenic enzymes in human placenta according to the gestational age. Mol. Med. Rep. 2019, 19, 3903–3911. [Google Scholar] [CrossRef] [PubMed]
- Walker, O.S.; Ragos, R.; Wong, M.K.; Adam, M.; Cheung, A.; Raha, S. Reactive oxygen species from mitochondria impacts trophoblast fusion and the production of endocrine hormones by syncytiotrophoblasts. PLoS ONE 2020, 15, e0229332. [Google Scholar] [CrossRef]
- Walker, O.S.; Ragos, R.; Gurm, H.; Lapierre, M.; May, L.L.; Raha, S. Delta-9-tetrahydrocannabinol disrupts mitochondrial function and attenuates syncytialization in human placental BeWo cells. Physiol. Rep. 2020, 8, e14476. [Google Scholar] [CrossRef]
- Zhu, H.; Ren, Q.; Yan, Z.; Xu, S.; Luo, J.; Wu, X.; Tang, C. Human HAND1 Inhibits the Conversion of Cholesterol to Steroids in Trophoblasts. J. Genet. Genom. 2021, 49, 350–363. [Google Scholar] [CrossRef]
- Ander, S.E.; Diamond, M.S.; Coyne, C.B. Immune responses at the maternal-fetal interface. Sci. Immunol. 2019, 4, eaat6114. [Google Scholar] [CrossRef]
- Jung, E.; Romero, R.; Yeo, L.; Gomez-Lopez, N.; Chaemsaithong, P.; Jaovisidha, A.; Gotsch, F.; Erez, O. The etiology of preeclampsia. Am. J. Obstet. Gynecol. 2022, 226, S844–S866. [Google Scholar] [CrossRef]
- Turbeville, H.R.; Sasser, J.M. Preeclampsia beyond pregnancy: Long-term consequences for mother and child. Am. J. Physiol. Renal Physiol. 2020, 318, F1315–F1326. [Google Scholar] [CrossRef]
- American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins–Obstetrics and the Society for Maternal-Fetal Medicine. ACOG Practice Bulletin No. 204: Fetal Growth Restriction. Obstet. Gynecol. 2019, 133, e97–e109. [Google Scholar] [CrossRef]
- Wilcox, A.J.; Cortese, M.; McConnaughey, D.R.; Moster, D.; Basso, O. The limits of small-for-gestational-age as a high-risk category. Eur. J. Epidemiol. 2021, 36, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Walani, S.R. Global burden of preterm birth. Int. J. Gynaecol. Obstet. 2020, 150, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Szmuilowicz, E.D.; Josefson, J.L.; Metzger, B.E. Gestational Diabetes Mellitus. Endocrinol. Metab. Clin. N. Am. 2019, 48, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Devall, A.J.; Papadopoulou, A.; Podesek, M.; Haas, D.M.; Price, M.J.; Coomarasamy, A.; Gallos, I.D. Progestogens for preventing miscarriage: A network meta-analysis. Cochrane Database Syst. Rev. 2021, 4, CD013792. [Google Scholar] [PubMed]
- Gonzalez, T.L.; Eisman, L.E.; Joshi, N.V.; Flowers, A.E.; Wu, D.; Wang, Y.; Santiskulvong, C.; Tang, J.; Buttle, R.A.; Sauro, E.; et al. High-throughput miRNA sequencing of the human placenta: Expression throughout gestation. Epigenomics 2021, 13, 995–1012. [Google Scholar] [CrossRef] [PubMed]
- Gillet, V.; Ouellet, A.; Stepanov, Y.; Rodosthenous, R.S.; Croft, E.K.; Brennan, K.; Abdelouahab, N.; Baccarelli, A.; Takser, L. miRNA Profiles in Extracellular Vesicles From Serum Early in Pregnancies Complicated by Gestational Diabetes Mellitus. J. Clin. Endocrinol. Metab. 2019, 104, 5157–5169. [Google Scholar] [CrossRef]
- Vaiman, D. Genes, epigenetics and miRNA regulation in the placenta. Placenta 2017, 52, 127–133. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Yoshida, T.; Asano, Y.; Ui-Tei, K. Modulation of MicroRNA Processing by Dicer via Its Associated dsRNA Binding Proteins. Non-Coding RNA 2021, 7, 57. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Rani, V.; Sengar, R.S. Biogenesis and mechanisms of microRNA-mediated gene regulation. Biotechnol. Bioeng. 2022, 119, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.P.; Peterson, K.R.; Slawson, C. O-GlcNAcylation and O-GlcNAc Cycling Regulate Gene Transcription: Emerging Roles in Cancer. Cancers 2021, 13, 1666. [Google Scholar] [CrossRef] [PubMed]
- Sheng, P.; Fields, C.; Aadland, K.; Wei, T.; Kolaczkowski, O.; Gu, T.; Kolaczkowski, B.; Xie, M. Dicer cleaves 5′-extended microRNA precursors originating from RNA polymerase II transcription start sites. Nucleic Acids Res. 2018, 46, 5737–5752. [Google Scholar] [CrossRef] [PubMed]
- Lauressergues, D.; Couzigou, J.M.; Clemente, H.S.; Martinez, Y.; Dunand, C.; Becard, G.; Combier, J.P. Primary transcripts of microRNAs encode regulatory peptides. Nature 2015, 520, 90–93. [Google Scholar] [CrossRef]
- Roshandel, E.; Noorazar, L.; Farhadihosseinabadi, B.; Mehdizadeh, M.; Kazemi, M.H.; Parkhideh, S. PI3 kinase signaling pathway in hematopoietic cancers: A glance in miRNA’s role. J. Clin. Lab. Anal. 2021, 35, e23725. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, A.; Seimiya, H. Tankyrase promotes primary precursor miRNA processing to precursor miRNA. Biochem. Biophys. Res. Commun. 2020, 522, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Kian, R.; Moradi, S.; Ghorbian, S. Role of components of microRNA machinery in carcinogenesis. Exp. Oncol. 2018, 40, 2–9. [Google Scholar] [CrossRef]
- Michlewski, G.; Caceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef]
- Clancy, J.W.; Zhang, Y.; Sheehan, C.; D’Souza-Schorey, C. An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat. Cell Biol. 2019, 21, 856–866. [Google Scholar] [CrossRef]
- Dandewad, V.; Vindu, A.; Joseph, J.; Seshadri, V. Import of human miRNA-RISC complex into Plasmodium falciparum and regulation of the parasite gene expression. J. Biosci. 2019, 44, 50. [Google Scholar] [CrossRef]
- Abedini, F.; Ebrahimi, M.; Hosseinkhani, H. Technology of RNA Interference in Advanced Medicine. Microrna 2018, 7, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Kern, F.; Krammes, L.; Danz, K.; Diener, C.; Kehl, T.; Kuchler, O.; Fehlmann, T.; Kahraman, M.; Rheinheimer, S.; Aparicio-Puerta, E.; et al. Validation of human microRNA target pathways enables evaluation of target prediction tools. Nucleic Acids Res. 2021, 49, 127–144. [Google Scholar] [CrossRef]
- Roos, D.; de Boer, M. Mutations in cis that affect mRNA synthesis, processing and translation. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166166. [Google Scholar] [CrossRef]
- Ezhilarasan, D. MicroRNA interplay between hepatic stellate cell quiescence and activation. Eur. J. Pharmacol. 2020, 885, 173507. [Google Scholar] [CrossRef]
- Chu, Y.W.; Chang, K.P.; Chen, C.W.; Liang, Y.T.; Soh, Z.T.; Hsieh, L.C. miRgo: Integrating various off-the-shelf tools for identification of microRNA-target interactions by heterogeneous features and a novel evaluation indicator. Sci. Rep. 2020, 10, 1466. [Google Scholar] [CrossRef]
- Moser, G.; Guettler, J.; Forstner, D.; Gauster, M. Maternal Platelets-Friend or Foe of the Human Placenta? Int. J. Mol. Sci. 2019, 20, 5639. [Google Scholar] [CrossRef]
- Knofler, M.; Haider, S.; Saleh, L.; Pollheimer, J.; Gamage, T.; James, J. Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell. Mol. Life Sci. 2019, 76, 3479–3496. [Google Scholar] [CrossRef] [Green Version]
- Zhai, J.; Xiao, Z.; Wang, Y.; Wang, H. Human embryonic development: From peri-implantation to gastrulation. Trends Cell Biol. 2022, 32, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.M.; Ezashi, T.; Schulz, L.C.; Sugimoto, J.; Schust, D.J.; Khan, T.; Zhou, J. Syncytins expressed in human placental trophoblast. Placenta 2021, 113, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M.; Chuva de Sousa Lopes, S.M. Emerging in vitro platforms and omics technologies for studying the endometrium and early embryo-maternal interface in humans. Placenta 2022, 125, 36–46. [Google Scholar] [CrossRef]
- Burton, G.J.; Jauniaux, E. Placentation in the Human and Higher Primates. Adv. Anat. Embryol. Cell Biol. 2021, 234, 223–254. [Google Scholar] [PubMed]
- Aplin, J.D.; Jones, C.J.P. Cell dynamics in human villous trophoblast. Hum. Reprod. Update 2021, 27, 904–922. [Google Scholar] [CrossRef]
- Sheridan, M.A.; Zhao, X.; Fernando, R.C.; Gardner, L.; Perez-Garcia, V.; Li, Q.; Marsh, S.G.E.; Hamilton, R.; Moffett, A.; Turco, M.Y. Characterization of primary models of human trophoblast. Development 2021, 148, dev199749. [Google Scholar] [CrossRef] [PubMed]
- Spiliopoulos, M.; Kuo, C.Y.; Eranki, A.; Jacobs, M.; Rossi, C.T.; Iqbal, S.N.; Fisher, J.P.; Fries, M.H.; Kim, P.C.W. Characterizing placental stiffness using ultrasound shear-wave elastography in healthy and preeclamptic pregnancies. Arch. Gynecol. Obstet. 2020, 302, 1103–1112. [Google Scholar] [CrossRef]
- Stenhouse, C.; Seo, H.; Wu, G.; Johnson, G.A.; Bazer, F.W. Insights into the Regulation of Implantation and Placentation in Humans, Rodents, Sheep, and Pigs. Adv. Exp. Med. Biol. 2022, 1354, 25–48. [Google Scholar]
- Cattini, P.A.; Jin, Y.; Jarmasz, J.S.; Noorjahan, N.; Bock, M.E. Obesity and regulation of human placental lactogen production in pregnancy. J. Neuroendocrinol. 2020, 32, e12859. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Sang, Y.; Li, D.J.; Du, M. Crosstalk Between Trophoblasts and Decidual Immune Cells: The Cornerstone of Maternal-Fetal Immunotolerance. Front. Immunol. 2021, 12, 642392. [Google Scholar] [CrossRef]
- Treissman, J.; Yuan, V.; Baltayeva, J.; Le, H.T.; Castellana, B.; Robinson, W.P.; Beristain, A.G. Low oxygen enhances trophoblast column growth by potentiating differentiation of the extravillous lineage and promoting LOX activity. Development 2020, 147, dev181263. [Google Scholar] [CrossRef] [PubMed]
- James, J.L.; Boss, A.L.; Sun, C.; Allerkamp, H.H.; Clark, A.R. From stem cells to spiral arteries: A journey through early placental development. Placenta 2022, 125, 68–77. [Google Scholar] [CrossRef]
- Albrecht, E.D.; Pepe, G.J. Regulation of Uterine Spiral Artery Remodeling: A Review. Reprod. Sci. 2020, 27, 1932–1942. [Google Scholar] [CrossRef]
- Haram, K.; Mortensen, J.H.; Myking, O.; Roald, B.; Magann, E.F.; Morrison, J.C. Early development of the human placenta and pregnancy complications. J. Matern. Fetal Neonatal. Med. 2020, 33, 3538–3545. [Google Scholar] [CrossRef] [PubMed]
- Mouillet, J.F.; Chu, T.; Nelson, D.M.; Mishima, T.; Sadovsky, Y. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J. 2010, 24, 2030–2039. [Google Scholar] [CrossRef] [PubMed]
- Hornakova, A.; Kolkova, Z.; Holubekova, V.; Loderer, D.; Lasabova, Z.; Biringer, K.; Halasova, E. Diagnostic Potential of MicroRNAs as Biomarkers in the Detection of Preeclampsia. Genet. Test. Mol. Biomark. 2020, 24, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Gunel, T.; Hosseini, M.K.; Gumusoglu, E.; Kisakesen, H.I.; Benian, A.; Aydinli, K. Expression profiling of maternal plasma and placenta microRNAs in preeclamptic pregnancies by microarray technology. Placenta 2017, 52, 77–85. [Google Scholar] [CrossRef]
- Wang, W.; Feng, L.; Zhang, H.; Hachy, S.; Satohisa, S.; Laurent, L.C.; Parast, M.; Zheng, J.; Chen, D.B. Preeclampsia up-regulates angiogenesis-associated microRNA (i.e., miR-17, -20a, and -20b) that target ephrin-B2 and EPHB4 in human placenta. J. Clin. Endocrinol. Metab. 2012, 97, E1051–E1059. [Google Scholar] [CrossRef]
- Stribling, D.; Lei, Y.; Guardia, C.M.; Li, L.; Fields, C.J.; Nowialis, P.; Opavsky, R.; Renne, R.; Xie, M. A noncanonical microRNA derived from the snaR-A noncoding RNA targets a metastasis inhibitor. RNA 2021, 27, 694–709. [Google Scholar] [CrossRef]
- Vashukova, E.S.; Glotov, A.S.; Fedotov, P.V.; Efimova, O.A.; Pakin, V.S.; Mozgovaya, E.V.; Pendina, A.A.; Tikhonov, A.V.; Koltsova, A.S.; Baranov, V.S. Placental microRNA expression in pregnancies complicated by superimposed preeclampsia on chronic hypertension. Mol. Med. Rep. 2016, 14, 22–32. [Google Scholar] [CrossRef]
- Pomin, V.H.; Wang, X. Synthetic Oligosaccharide Libraries and Microarray Technology: A Powerful Combination for the Success of Current Glycosaminoglycan Interactomics. ChemMedChem 2018, 13, 648–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaylak, B.; Akgul, B. Experimental MicroRNA Detection Methods. Methods Mol. Biol. 2022, 2257, 33–55. [Google Scholar] [PubMed]
- Huang, Y.; Ren, H.T.; Xiong, J.L.; Gao, X.C.; Sun, X.H. Identification and characterization of known and novel microRNAs in three tissues of Chinese giant salamander base on deep sequencing approach. Genomics 2017, 109, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Hayder, H.; O’Brien, J.; Nadeem, U.; Peng, C. MicroRNAs: Crucial regulators of placental development. Reproduction 2018, 155, R259–R271. [Google Scholar] [CrossRef]
- Morales-Prieto, D.M.; Ospina-Prieto, S.; Schmidt, A.; Chaiwangyen, W.; Markert, U.R. Elsevier Trophoblast Research Award Lecture: Origin, evolution and future of placenta miRNAs. Placenta 2014, 35, S39–S45. [Google Scholar] [CrossRef]
- Morales-Prieto, D.M.; Chaiwangyen, W.; Ospina-Prieto, S.; Schneider, U.; Herrmann, J.; Gruhn, B.; Markert, U.R. MicroRNA expression profiles of trophoblastic cells. Placenta 2012, 33, 725–734. [Google Scholar] [CrossRef]
- Chen, P.S.; Chiu, W.T.; Hsu, P.L.; Lin, S.C.; Peng, I.C.; Wang, C.Y.; Tsai, S.J. Pathophysiological implications of hypoxia in human diseases. J. Biomed. Sci. 2020, 27, 63. [Google Scholar] [CrossRef]
- Hu, X.Q.; Zhang, L. Hypoxia and Mitochondrial Dysfunction in Pregnancy Complications. Antioxidants 2021, 10, 405. [Google Scholar] [CrossRef]
- Maslen, C.L. Recent Advances in Placenta-Heart Interactions. Front. Physiol. 2018, 9, 735. [Google Scholar] [CrossRef]
- Burton, G.J.; Cindrova-Davies, T.; Yung, H.W.; Jauniaux, E. Hypoxia and Reproductive Health: Oxygen and development of the human placenta. Reproduction 2021, 161, F53–F65. [Google Scholar] [CrossRef]
- Jaszczuk, I.; Koczkodaj, D.; Kondracka, A.; Kwasniewska, A.; Winkler, I.; Filip, A. The role of miRNA-210 in pre-eclampsia development. Ann. Med. 2022, 54, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Chen, X.; Wang, L.; Chen, F.; Cen, H.; Shi, L. Hypoxia-induced microRNA-141 regulates trophoblast apoptosis, invasion, and vascularization by blocking CXCL12beta/CXCR2/4 signal transduction. Biomed. Pharmacother. 2019, 116, 108836. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Du, H.; Han, B.; Xia, G.; Shi, X.; Zhang, F.; Fu, Q.; Zhang, T. Hypoxia-inducible microRNA-218 inhibits trophoblast invasion by targeting LASP1: Implications for preeclampsia development. Int. J. Biochem. Cell Biol. 2017, 87, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Addo, K.A.; Palakodety, N.; Hartwell, H.J.; Tingare, A.; Fry, R.C. Placental microRNAs: Responders to environmental chemicals and mediators of pathophysiology of the human placenta. Toxicol. Rep. 2020, 7, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Hadlich, F.; Abbas, M.W.; Iqbal, M.A.; Tesfaye, D.; Bouma, G.J.; Winger, Q.A.; Ponsuksili, S. MicroRNA-mRNA Networks in Pregnancy Complications: A Comprehensive Downstream Analysis of Potential Biomarkers. Int. J. Mol. Sci. 2021, 22, 2313. [Google Scholar] [CrossRef]
- Eaves, L.A.; Phookphan, P.; Rager, J.E.; Bangma, J.; Santos, H.P., Jr.; Smeester, L.; O’Shea, T.M.; Fry, R.C. A role for microRNAs in the epigenetic control of sexually dimorphic gene expression in the human placenta. Epigenomics 2020, 12, 1543–1558. [Google Scholar] [CrossRef]
- Zhong, J.; Baccarelli, A.A.; Mansur, A.; Adir, M.; Nahum, R.; Hauser, R.; Bollati, V.; Racowsky, C.; Machtinger, R. Maternal Phthalate and Personal Care Products Exposure Alters Extracellular Placental miRNA Profile in Twin Pregnancies. Reprod. Sci. 2019, 26, 289–294. [Google Scholar] [CrossRef]
- Meruvu, S.; Zhang, J.; Choudhury, M. Mono-(2-ethylhexyl) Phthalate Increases Oxidative Stress Responsive miRNAs in First Trimester Placental Cell Line HTR8/SVneo. Chem. Res. Toxicol. 2016, 29, 430–435. [Google Scholar] [CrossRef]
- Meruvu, S.; Zhang, J.; Bedi, Y.S.; Choudhury, M. Mono-(2-ethylhexyl) phthalate induces apoptosis through miR-16 in human first trimester placental cell line HTR-8/SVneo. Toxicol. In Vitro 2016, 31, 35–42. [Google Scholar] [CrossRef]
- Avissar-Whiting, M.; Veiga, K.R.; Uhl, K.M.; Maccani, M.A.; Gagne, L.A.; Moen, E.L.; Marsit, C.J. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod. Toxicol. 2010, 29, 401–406. [Google Scholar] [CrossRef]
- LaRocca, J.; Binder, A.M.; McElrath, T.F.; Michels, K.B. First-Trimester Urine Concentrations of Phthalate Metabolites and Phenols and Placenta miRNA Expression in a Cohort of U.S. Women. Environ. Health Perspect. 2016, 124, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Liu, Y.; Dong, S. Emerging roles of long non-coding RNAs in the toxicology of environmental chemicals. J. Appl. Toxicol. 2018, 38, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Noguer-Dance, M.; Abu-Amero, S.; Al-Khtib, M.; Lefevre, A.; Coullin, P.; Moore, G.E.; Cavaille, J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum. Mol. Genet. 2010, 19, 3566–3582. [Google Scholar] [CrossRef] [PubMed]
- Prats-Puig, A.; Xargay-Torrent, S.; Carreras-Badosa, G.; Mas-Pares, B.; Bassols, J.; Petry, C.J.; Girardot, M.; Francis, D.E.Z.; Ibanez, L.; Dunger, D.B.; et al. Methylation of the C19MC microRNA locus in the placenta: Association with maternal and chilhood body size. Int. J. Obes. 2020, 44, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.; Bamba, C.; Chopra, S.; Mandal, K. Role of miRNA polymorphism in recurrent pregnancy loss: A systematic review and meta-analysis. Biomark Med. 2022, 16, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Ezat, S.A.; Haji, A.I. Study of association between different microRNA variants and the risk of idiopathic recurrent pregnancy loss. Arch. Gynecol. Obstet. 2022, 306, 1281–1286. [Google Scholar] [CrossRef]
- Inno, R.; Kikas, T.; Lillepea, K.; Laan, M. Coordinated Expressional Landscape of the Human Placental miRNome and Transcriptome. Front. Cell Dev. Biol. 2021, 9, 697947. [Google Scholar] [CrossRef]
- Mouillet, J.F.; Goff, J.; Sadovsky, E.; Sun, H.; Parks, T.; Chu, T.; Sadovsky, Y. Transgenic expression of human C19MC miRNAs impacts placental morphogenesis. Placenta 2020, 101, 208–214. [Google Scholar] [CrossRef]
- Fu, G.; Ye, G.; Nadeem, L.; Ji, L.; Manchanda, T.; Wang, Y.; Zhao, Y.; Qiao, J.; Wang, Y.L.; Lye, S.; et al. MicroRNA-376c impairs transforming growth factor-beta and nodal signaling to promote trophoblast cell proliferation and invasion. Hypertension 2013, 61, 864–872. [Google Scholar] [CrossRef]
- Luo, L.; Ye, G.; Nadeem, L.; Fu, G.; Yang, B.B.; Honarparvar, E.; Dunk, C.; Lye, S.; Peng, C. MicroRNA-378a-5p promotes trophoblast cell survival, migration and invasion by targeting Nodal. J. Cell Sci. 2012, 125, 3124–3132. [Google Scholar]
- Gottlieb, A.; Flor, I.; Nimzyk, R.; Burchardt, L.; Helmke, B.; Langenbuch, M.; Spiekermann, M.; Feidicker, S.; Bullerdiek, J. The expression of miRNA encoded by C19MC and miR-371-3 strongly varies among individual placentas but does not differ between spontaneous and induced abortions. Protoplasma 2021, 258, 209–218. [Google Scholar] [CrossRef]
- Forbes, K.; Farrokhnia, F.; Aplin, J.D.; Westwood, M. Dicer-dependent miRNAs provide an endogenous restraint on cytotrophoblast proliferation. Placenta 2012, 33, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, X.; He, Z.; Xiong, Y.; Fang, Q. miRNA-210-3p regulates trophoblast proliferation and invasiveness through fibroblast growth factor 1 in selective intrauterine growth restriction. J. Cell. Mol. Med. 2019, 23, 4422–4433. [Google Scholar] [CrossRef]
- Wang, D.; Na, Q.; Song, G.Y.; Wang, L. Human umbilical cord mesenchymal stem cell-derived exosome-mediated transfer of microRNA-133b boosts trophoblast cell proliferation, migration and invasion in preeclampsia by restricting SGK1. Cell Cycle 2020, 19, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, Y.; Lu, H.; Wang, H.; Shi, X.; Shao, X.; Li, Y.X.; Zhao, Y.; Wang, Y.L. miR-518b Enhances Human Trophoblast Cell Proliferation Through Targeting Rap1b and Activating Ras-MAPK Signal. Front. Endocrinol. 2018, 9, 100. [Google Scholar] [CrossRef]
- Dai, Y.; Qiu, Z.; Diao, Z.; Shen, L.; Xue, P.; Sun, H.; Hu, Y. MicroRNA-155 inhibits proliferation and migration of human extravillous trophoblast derived HTR-8/SVneo cells via down-regulating cyclin D1. Placenta 2012, 33, 824–829. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; He, Z.; Cai, J.; Huang, L.; Zhu, H.; Luo, Y. Potential role of microRNA-424 in regulating ERRgamma to suppress trophoblast proliferation and invasion in fetal growth restriction. Placenta 2019, 83, 57–62. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, J.; Li, M.Q.; Xu, J.; Zhang, J.P.; Jin, L.P. MicroRNA-184 promotes apoptosis of trophoblast cells via targeting WIG1 and induces early spontaneous abortion. Cell Death Dis. 2019, 10, 223. [Google Scholar] [CrossRef]
- Liu, H.N.; Tang, X.M.; Wang, X.Q.; Gao, J.; Li, N.; Wang, Y.Y.; Xia, H.F. MiR-93 Inhibits Trophoblast Cell Proliferation and Promotes Cell Apoptosis by Targeting BCL2L2 in Recurrent Spontaneous Abortion. Reprod. Sci. 2020, 27, 152–162. [Google Scholar] [CrossRef]
- Matsubara, K.; Matsubara, Y.; Uchikura, Y.; Takagi, K.; Yano, A.; Sugiyama, T. HMGA1 Is a Potential Driver of Preeclampsia Pathogenesis by Interference with Extravillous Trophoblasts Invasion. Biomolecules 2021, 11, 822. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, A.; Shima, T.; Aoki, A.; Kawaguchi, M.; Yasuda, I.; Tsuda, S.; Yoneda, S.; Yamaki-Ushijima, A.; Cheng, S.B.; Sharma, S.; et al. Molecular and immunological developments in placentas. Hum. Immunol. 2021, 82, 317–324. [Google Scholar] [CrossRef]
- Sun, M.; Chen, H.; Liu, J.; Tong, C.; Meng, T. MicroRNA-34a inhibits human trophoblast cell invasion by targeting MYC. BMC Cell Biol. 2015, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Chen, A.; Yang, H.; Hong, L. MicroRNA-27a inhibits trophoblast cell migration and invasion by targeting SMAD2: Potential role in preeclampsia. Exp. Ther. Med. 2020, 20, 2262–2269. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Deng, M.; Wang, Q. MiRNA-320a inhibits trophoblast cell invasion by targeting estrogen-related receptor-gamma. J. Obstet. Gynaecol. Res. 2018, 44, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Meng, T. MicroRNA-431 affects trophoblast migration and invasion by targeting ZEB1 in preeclampsia. Gene 2019, 683, 225–232. [Google Scholar] [CrossRef]
- Niu, Z.R.; Han, T.; Sun, X.L.; Luan, L.X.; Gou, W.L.; Zhu, X.M. MicroRNA-30a-3p is overexpressed in the placentas of patients with preeclampsia and affects trophoblast invasion and apoptosis by its effects on IGF-1. Am. J. Obstet. Gynecol. 2018, 218, 249.e1–249.e12. [Google Scholar] [CrossRef]
- Mandl, M.; Haas, J.; Bischof, P.; Nohammer, G.; Desoye, G. Serum-dependent effects of IGF-I and insulin on proliferation and invasion of human first trimester trophoblast cell models. Histochem. Cell Biol. 2002, 117, 391–399. [Google Scholar] [CrossRef]
- Shih, J.C.; Lin, H.H.; Hsiao, A.C.; Su, Y.T.; Tsai, S.; Chien, C.L.; Kung, H.N. Unveiling the role of microRNA-7 in linking TGF-beta-Smad-mediated epithelial-mesenchymal transition with negative regulation of trophoblast invasion. FASEB J. 2019, 33, 6281–6295. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, L.; Liu, T.; Guan, H. MicroRNA-204 suppresses trophoblast-like cell invasion by targeting matrix metalloproteinase-9. Biochem. Biophys. Res. Commun. 2015, 463, 285–291. [Google Scholar] [CrossRef]
- Tamaru, S.; Mizuno, Y.; Tochigi, H.; Kajihara, T.; Okazaki, Y.; Okagaki, R.; Kamei, Y.; Ishihara, O.; Itakura, A. MicroRNA-135b suppresses extravillous trophoblast-derived HTR-8/SVneo cell invasion by directly down regulating CXCL12 under low oxygen conditions. Biochem. Biophys. Res. Commun. 2015, 461, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Jiang, L.; Gu, X.; Huang, S.; Pang, J.; Wu, Y.; Yin, J.; Wang, J. SIRT3 deficiency is resistant to autophagy-dependent ferroptosis by inhibiting the AMPK/mTOR pathway and promoting GPX4 levels. J. Cell. Physiol. 2020, 235, 8839–8851. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Chen, Y.; Long, Y.; Yu, J.; Li, M. Tumor necrosis factor-alpha suppresses the invasion of HTR-8/SVneo trophoblast cells through microRNA-145-5p-mediated downregulation of Cyr61. Life Sci. 2018, 209, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Xu, S.; Li, J.; Yao, Y.; Tang, C. MicroRNA-3935 promotes human trophoblast cell epithelial-mesenchymal transition through tumor necrosis factor receptor-associated factor 6/regulator of G protein signaling 2 axis. Reprod. Biol. Endocrinol. 2021, 19, 134. [Google Scholar] [CrossRef]
- Wu, M.; Zhao, Y.; Li, L.; Wang, G.; Xing, L. Exosomal microRNA302a promotes trophoblast migration and proliferation, and represses angiogenesis by regulating the expression levels of VEGFA in preeclampsia. Mol. Med. Rep. 2021, 24, 864. [Google Scholar] [CrossRef]
- Rana, S.; Lemoine, E.; Granger, J.P.; Karumanchi, S.A. Preeclampsia: Pathophysiology, Challenges, and Perspectives. Circ. Res. 2019, 124, 1094–1112. [Google Scholar] [CrossRef]
- Armaly, Z.; Jadaon, J.E.; Jabbour, A.; Abassi, Z.A. Preeclampsia: Novel Mechanisms and Potential Therapeutic Approaches. Front. Physiol. 2018, 9, 973. [Google Scholar] [CrossRef]
- Yang, H.L.; Zhang, H.Z.; Meng, F.R.; Han, S.Y.; Zhang, M. Differential expression of microRNA-411 and 376c is associated with hypertension in pregnancy. Braz. J. Med. Biol. Res. 2019, 52, e7546. [Google Scholar] [CrossRef]
- Akehurst, C.; Small, H.Y.; Sharafetdinova, L.; Forrest, R.; Beattie, W.; Brown, C.E.; Robinson, S.W.; McClure, J.D.; Work, L.M.; Carty, D.M.; et al. Differential expression of microRNA-206 and its target genes in preeclampsia. J. Hypertens. 2015, 33, 2068–2074. [Google Scholar] [CrossRef]
- Gunel, T.; Kamali, N.; Hosseini, M.K.; Gumusoglu, E.; Benian, A.; Aydinli, K. Regulatory effect of miR-195 in the placental dysfunction of preeclampsia. J. Matern. Fetal Neonatal Med. 2020, 33, 901–908. [Google Scholar] [CrossRef]
- Muralimanoharan, S.; Maloyan, A.; Mele, J.; Guo, C.; Myatt, L.G.; Myatt, L. MIR-210 modulates mitochondrial respiration in placenta with preeclampsia. Placenta 2012, 33, 816–823. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Wang, H.; Wang, J.; Zhang, Y.; Wang, Y.; Pan, Z.; Luo, S. Aberrantly up-regulated miR-20a in pre-eclampsic placenta compromised the proliferative and invasive behaviors of trophoblast cells by targeting forkhead box protein A1. Int. J. Biol. Sci. 2014, 10, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Li, H.; Xu, H.; Huo, G.; Yao, Y. MicroRNA-20b inhibits trophoblast cell migration and invasion by targeting MMP-2. Int. J. Clin. Exp. Pathol. 2017, 10, 10901–10909. [Google Scholar] [PubMed]
- Du, J.; Ji, Q.; Dong, L.; Meng, Y.; Xin, G. HDAC4 Knockdown Induces Preeclampsia Cell Autophagy and Apoptosis by miR-29b. Reprod. Sci. 2021, 28, 334–342. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhao, L.; Wang, X.; Lian, F.; Cai, Y. Ligustrazine-induced microRNA-16-5p inhibition alleviates preeclampsia through IGF-2. Reproduction 2020, 160, 905–917. [Google Scholar] [CrossRef]
- Beards, F.; Jones, L.E.; Charnock, J.; Forbes, K.; Harris, L.K. Placental Homing Peptide-microRNA Inhibitor Conjugates for Targeted Enhancement of Intrinsic Placental Growth Signaling. Theranostics 2017, 7, 2940–2955. [Google Scholar] [CrossRef]
- Ospina-Prieto, S.; Chaiwangyen, W.; Herrmann, J.; Groten, T.; Schleussner, E.; Markert, U.R.; Morales-Prieto, D.M. MicroRNA-141 is upregulated in preeclamptic placentae and regulates trophoblast invasion and intercellular communication. Transl. Res. 2016, 172, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Hou, B.; Shan, L.; Sun, X.; Wang, L. Aberrantly up-regulated miR-142-3p inhibited the proliferation and invasion of trophoblast cells by regulating FOXM1. Placenta 2021, 104, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wang, D.; Lu, J.; Zhou, X. MiR-125b participates in the occurrence of preeclampsia by regulating the migration and invasion of extravillous trophoblastic cells through STAT3 signaling pathway. J. Recept. Signal. Transduct. Res. 2021, 41, 202–208. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X. MiR-200b-3p is upregulated in the placental tissues from patients with preeclampsia and promotes the development of preeclampsia via targeting profilin 2. Cell Cycle 2022, 21, 1945–1957. [Google Scholar] [CrossRef]
- Lu, T.M.; Lu, W.; Zhao, L.J. MicroRNA-137 Affects Proliferation and Migration of Placenta Trophoblast Cells in Preeclampsia by Targeting ERRalpha. Reprod. Sci. 2017, 24, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Williams, J., III; Brown, J.; Wang, E.T.; Lee, B.; Gonzalez, T.L.; Cui, J.; Goodarzi, M.O.; Pisarska, M.D. Up-regulation of microRNA-202-3p in first trimester placenta of pregnancies destined to develop severe preeclampsia, a pilot study. Pregnancy Hypertens. 2017, 10, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Song, H.; Xie, C.; Zheng, W.; Ma, H.; Xin, D.; Zhan, J.; Yuan, X.; Chen, A.; Tao, J.; et al. miR-146a-5p-mediated suppression on trophoblast cell progression and epithelial-mesenchymal transition in preeclampsia. Biol. Res. 2021, 54, 30. [Google Scholar] [CrossRef] [PubMed]
- Nizyaeva, N.V.; Kulikova, G.V.; Nagovitsyna, M.N.; Kan, N.E.; Prozorovskaya, K.N.; Shchegolev, A.I.; Sukhikh, G.T. Expression of MicroRNA-146a and MicroRNA-155 in Placental Villi in Early- and Late-Onset Preeclampsia. Bull. Exp. Biol. Med. 2017, 163, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Anton, L.; Olarerin-George, A.O.; Hogenesch, J.B.; Elovitz, M.A. Placental expression of miR-517a/b and miR-517c contributes to trophoblast dysfunction and preeclampsia. PLoS ONE 2015, 10, e0122707. [Google Scholar] [CrossRef]
- Kumar, P.; Luo, Y.; Tudela, C.; Alexander, J.M.; Mendelson, C.R. The c-Myc-regulated microRNA-17~92 (miR-17~92) and miR-106a~363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol. Cell Biol. 2013, 33, 1782–1796. [Google Scholar] [CrossRef]
- Baker, A.H.; Delles, C. Is microRNA-376c a biomarker or mediator of preeclampsia? Hypertension 2013, 61, 767–769. [Google Scholar] [CrossRef]
- Sandrim, V.C.; Eleuterio, N.; Pilan, E.; Tanus-Santos, J.E.; Fernandes, K.; Cavalli, R. Plasma levels of increased miR-195-5p correlates with the sFLT-1 levels in preeclampsia. Hypertens. Pregnancy 2016, 35, 150–158. [Google Scholar] [CrossRef]
- Jiang, L.; Li, M.; Gan, H.; Yang, L.; Lin, F.; Hu, M. miR-335 targets CRIM1 to promote the proliferation and inhibit the apoptosis of placental trophoblast cells in preeclamptic rats. Am. J. Transl. Res. 2021, 13, 4676–4685. [Google Scholar]
- Yan, T.; Liu, Y.; Cui, K.; Hu, B.; Wang, F.; Zou, L. MicroRNA-126 regulates EPCs function: Implications for a role of miR-126 in preeclampsia. J. Cell Biochem. 2013, 114, 2148–2159. [Google Scholar] [CrossRef]
- Gu, Y.; Meng, J.; Zuo, C.; Wang, S.; Li, H.; Zhao, S.; Huang, T.; Wang, X.; Yan, J. Downregulation of MicroRNA-125a in Placenta Accreta Spectrum Disorders Contributes Antiapoptosis of Implantation Site Intermediate Trophoblasts by Targeting MCL1. Reprod. Sci. 2019, 26, 1582–1589. [Google Scholar] [CrossRef]
- Li, H.; Ouyang, Y.; Sadovsky, E.; Parks, W.T.; Chu, T.; Sadovsky, Y. Unique microRNA Signals in Plasma Exosomes from Pregnancies Complicated by Preeclampsia. Hypertension 2020, 75, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Salomon, C.; Guanzon, D.; Scholz-Romero, K.; Longo, S.; Correa, P.; Illanes, S.E.; Rice, G.E. Placental Exosomes as Early Biomarker of Preeclampsia: Potential Role of Exosomal MicroRNAs Across Gestation. J. Clin. Endocrinol. Metab. 2017, 102, 3182–3194. [Google Scholar] [CrossRef]
- Shen, L.; Li, Y.; Li, R.; Diao, Z.; Yany, M.; Wu, M.; Sun, H.; Yan, G.; Hu, Y. Placentaassociated serum exosomal miR155 derived from patients with preeclampsia inhibits eNOS expression in human umbilical vein endothelial cells. Int. J. Mol. Med. 2018, 41, 1731–1739. [Google Scholar]
- Motawi, T.M.K.; Sabry, D.; Maurice, N.W.; Rizk, S.M. Role of mesenchymal stem cells exosomes derived microRNAs; miR-136, miR-494 and miR-495 in pre-eclampsia diagnosis and evaluation. Arch. Biochem. Biophys. 2018, 659, 13–21. [Google Scholar] [CrossRef]
- Rokni, M.; Salimi, S.; Sohrabi, T.; Asghari, S.; Teimoori, B.; Saravani, M. Association between miRNA-152 polymorphism and risk of preeclampsia susceptibility. Arch. Gynecol. Obstet. 2019, 299, 475–480. [Google Scholar] [CrossRef]
- Maharaj, N.R.; Ramkaran, P.; Pillay, S.; Chuturgoon, A.A. MicroRNA-146a rs2910164 is associated with severe preeclampsia in Black South African women on HAART. BMC Genet. 2017, 18, 5. [Google Scholar] [CrossRef] [PubMed]
- Eskandari, F.; Rezaei, M.; Mohammadpour-Gharehbagh, A.; Teimoori, B.; Yaghmaei, M.; Narooei-Nejad, M.; Salimi, S. The association of pri-miRNA- 26a1 rs7372209 polymorphism and Preeclampsia susceptibility. Clin. Exp. Hypertens. 2019, 41, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, S.E.; Shaker, O.G.; Abdelwahed, M.Y.; Ahmed, N.A.; Abdelhameed, H.G.; Bosilah, A.H.; Mohammed, S.R. Association of MicroRNA-155rs767649 Polymorphism with Susceptibility to Preeclampsia. Int. J. Mol. Cell. Med. 2019, 8, 247–257. [Google Scholar]
- Abo-Elmatty, D.M.; Mehanna, E.T. MIR146A rs2910164 (G/C) Polymorphism is Associated with Incidence of Preeclampsia in Gestational Diabetes Patients. Biochem. Genet. 2019, 57, 222–233. [Google Scholar] [CrossRef]
- Meng, M.; Cheng, Y.K.Y.; Wu, L.; Chaemsaithong, P.; Leung, M.B.W.; Chim, S.S.C.; Sahota, D.S.; Li, W.; Poon, L.C.Y.; Wang, C.C.; et al. Whole genome miRNA profiling revealed miR-199a as potential placental pathogenesis of selective fetal growth restriction in monochorionic twin pregnancies. Placenta 2020, 92, 44–53. [Google Scholar] [CrossRef]
- Huang, L.; Shen, Z.; Xu, Q.; Huang, X.; Chen, Q.; Li, D. Increased levels of microRNA-424 are associated with the pathogenesis of fetal growth restriction. Placenta 2013, 34, 624–627. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Na, Q.; Song, W.W.; Song, G.Y. Altered Expression of miR-518b and miR-519a in the placenta is associated with low fetal birth weight. Am. J. Perinatol. 2014, 31, 729–734. [Google Scholar] [CrossRef]
- Higashijima, A.; Miura, K.; Mishima, H.; Kinoshita, A.; Jo, O.; Abe, S.; Hasegawa, Y.; Miura, S.; Yamasaki, K.; Yoshida, A.; et al. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat. Diagn. 2013, 33, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Maccani, M.A.; Padbury, J.F.; Marsit, C.J. miR-16 and miR-21 expression in the placenta is associated with fetal growth. PLoS ONE 2011, 6, e21210. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.L.; Zhang, L.; Li, J.; Tian, S.; Lv, X.D.; Wang, X.Q.; Su, X.; Li, Y.; Hu, Y.; Ma, X.; et al. Up-regulation of miR-98 and unraveling regulatory mechanisms in gestational diabetes mellitus. Sci. Rep. 2016, 6, 32268. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, T.; Shi, Z.; Ding, H.; Ling, X. MicroRNA-518d regulates PPARalpha protein expression in the placentas of females with gestational diabetes mellitus. Mol. Med. Rep. 2014, 9, 2085–2090. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, T.; Sun, D.; Cheng, G.; Ren, H.; Hong, H.; Chen, L.; Jiao, X.; Du, Y.; Zou, Y.; et al. Diagnostic value of dysregulated microribonucleic acids in the placenta and circulating exosomes in gestational diabetes mellitus. J. Diabetes Investig. 2021, 12, 1490–1500. [Google Scholar] [CrossRef]
- Sebastiani, G.; Guarino, E.; Grieco, G.E.; Formichi, C.; Delli Poggi, C.; Ceccarelli, E.; Dotta, F. Circulating microRNA (miRNA) Expression Profiling in Plasma of Patients with Gestational Diabetes Mellitus Reveals Upregulation of miRNA miR-330-3p. Front. Endocrinol. 2017, 8, 345. [Google Scholar] [CrossRef]
- Sun, D.G.; Tian, S.; Zhang, L.; Hu, Y.; Guan, C.Y.; Ma, X.; Xia, H.F. The miRNA-29b Is Downregulated in Placenta During Gestational Diabetes Mellitus and May Alter Placenta Development by Regulating Trophoblast Migration and Invasion Through a HIF3A-Dependent Mechanism. Front. Endocrinol. 2020, 11, 169. [Google Scholar] [CrossRef]
- Li, L.; Wang, S.; Li, H.; Wan, J.; Zhou, Q.; Zhou, Y.; Zhang, C. microRNA-96 protects pancreatic beta-cell function by targeting PAK1 in gestational diabetes mellitus. Biofactors 2018, 44, 539–547. [Google Scholar] [CrossRef]
- Peng, H.Y.; Li, M.Q.; Li, H.P. High glucose suppresses the viability and proliferation of HTR8/SVneo cells through regulation of the miR137/PRKAA1/IL6 axis. Int. J. Mol. Med. 2018, 42, 799–810. [Google Scholar] [PubMed]
- Herrera-Van Oostdam, A.S.; Toro-Ortiz, J.C.; Lopez, J.A.; Noyola, D.E.; Garcia-Lopez, D.A.; Duran-Figueroa, N.V.; Martinez-Martinez, E.; Portales-Perez, D.P.; Salgado-Bustamante, M.; López Hernández, Y. Placental exosomes isolated from urine of patients with gestational diabetes exhibit a differential profile expression of microRNAs across gestation. Int. J. Mol. Med. 2020, 46, 546–560. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Shen, W.W.; Cao, X.M.; Ding, W.Y.; Yan, L.P.; Gao, L.J.; Li, X.L.; Zhong, T.Y. Novel mechanism of miRNA-365-regulated trophoblast apoptosis in recurrent miscarriage. J. Cell. Mol. Med. 2017, 21, 2412–2425. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, X.; Chang, Y.; Wang, Y.; Xu, X.; Guo, Y.; Cui, H. Inhibition of microRNA-149 protects against recurrent miscarriage through upregulating RUNX2 and activation of the PTEN/Akt signaling pathway. J. Obstet. Gynaecol. Res. 2020, 46, 2534–2546. [Google Scholar] [CrossRef]
- Tang, H.; Pan, L.; Xiong, Y.; Wang, L.; Cui, Y.; Liu, J.; Tang, L. Down-regulation of the Sp1 transcription factor by an increase of microRNA-4497 in human placenta is associated with early recurrent miscarriage. Reprod. Biol. Endocrinol. 2021, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tian, P.; Xu, H. MicroRNA-155-5p regulates survival of human decidua stromal cells through NF-kappaB in recurrent miscarriage. Reprod. Biol. 2021, 21, 100510. [Google Scholar] [CrossRef]
- Montenegro, D.; Romero, R.; Kim, S.S.; Tarca, A.L.; Draghici, S.; Kusanovic, J.P.; Kim, J.S.; Lee, D.C.; Erez, O.; Gotsch, F.; et al. Expression patterns of microRNAs in the chorioamniotic membranes: A role for microRNAs in human pregnancy and parturition. J. Pathol. 2009, 217, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Marttila, S.; Rovio, S.; Mishra, P.P.; Seppala, I.; Lyytikainen, L.P.; Juonala, M.; Waldenberger, M.; Oksala, N.; Ala-Korpela, M.; Harville, E.; et al. Adulthood blood levels of hsa-miR-29b-3p associate with preterm birth and adult metabolic and cognitive health. Sci. Rep. 2021, 11, 9203. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Brkic, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef]
- Naidoo, P.; Naidoo, R.N.; Ramkaran, P.; Muttoo, S.; Asharam, K.; Chuturgoon, A.A. Maternal miRNA-146a G/C rs2910164 variation, HIV/AIDS and nitrogen oxide pollution exposure collectively affects foetal growth. Hum. Exp. Toxicol. 2019, 38, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Coustan, D.R. Gestational diabetes mellitus. Clin. Chem. 2013, 59, 1310–1321. [Google Scholar] [CrossRef]
- Yang, X.; Wu, N. MicroRNAs and Exosomal microRNAs May Be Possible Targets to Investigate in Gestational Diabetes Mellitus. Diabetes Metab. Syndr. Obes. 2022, 15, 321–330. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, R.; Liu, C.; Ma, C.; Chen, X.; Yang, J.; Sun, D. MicroRNA single-nucleotide polymorphisms and diabetes mellitus: A comprehensive review. Clin. Genet. 2019, 95, 451–461. [Google Scholar] [CrossRef]
- Wang, X.; Li, W.; Ma, L.; Ping, F.; Liu, J.; Wu, X.; Mao, J.; Wang, X.; Nie, M. Investigation of miRNA-binding site variants and risk of gestational diabetes mellitus in Chinese pregnant women. Acta Diabetol. 2017, 54, 309–316. [Google Scholar] [CrossRef]
- Wang, X.; Li, W.; Ma, L.; Gao, J.; Liu, J.; Ping, F.; Nie, M. Association study of the miRNA-binding site polymorphisms of CDKN2A/B genes with gestational diabetes mellitus susceptibility. Acta Diabetol. 2015, 52, 951–958. [Google Scholar] [CrossRef]
- Hakimian, M.; Ghorbian, S. Negative associations between the has-miR-27a and hsa-miR-125a gene variations and prostate cancer susceptibility. Mol. Biol. Rep. 2020, 47, 4209–4214. [Google Scholar] [CrossRef]
- Menon, R.; Debnath, C.; Lai, A.; Guanzon, D.; Bhaatnagar, S.; Kshetrapal, P.K.; Sheller-Miller, S.; Salomon, C.; Garbhini Study, T. Circulating Exosomal miRNA Profile During Term and Preterm Birth Pregnancies: A Longitudinal Study. Endocrinology 2019, 160, 249–275. [Google Scholar] [CrossRef]
- Menon, R.; Shahin, H. Extracellular vesicles in spontaneous preterm birth. Am. J. Reprod. Immunol. 2021, 85, e13353. [Google Scholar] [CrossRef]
- Jeong, H.R.; Han, J.A.; Kim, H.; Lee, H.J.; Shim, Y.S.; Kang, M.J.; Yoon, J.S.; Ryu, S.; Hwang, I.T. Exosomal miRNA Profile in Small-for-Gestational-Age Children: A Potential Biomarker for Catch-Up Growth. Genes 2022, 13, 938. [Google Scholar] [CrossRef]
- Rah, H.; Chung, K.W.; Ko, K.H.; Kim, E.S.; Kim, J.O.; Sakong, J.H.; Kim, J.H.; Lee, W.S.; Kim, N.K. miR-27a and miR-449b polymorphisms associated with a risk of idiopathic recurrent pregnancy loss. PLoS ONE 2017, 12, e0177160. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Kim, E.S.; Ahn, E.H.; Kim, J.O.; An, H.J.; Kim, J.H.; Lee, Y.; Lee, W.S.; Kim, Y.R.; Kim, N.K. The microRNApolymorphisms inmiR-150 and miR-1179 are associated with risk of idiopathic recurrent pregnancy loss. Reprod. Biomed. Online 2019, 39, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Chen, Y.; Dai, J.; Wang, B.; Liu, M.; Wang, Y.; Tao, J.; Li, H. Methylenetetrahydrofolate reductase polymorphisms at 3′-untranslated region are associated with susceptibility to preterm birth. Transl. Pediatr. 2015, 4, 57–62. [Google Scholar] [PubMed]
- Silva, L.R.; Melo, A.S.; Salomao, K.B.; Mazin, S.C.; Tone, L.G.; Cardoso, V.C.; Dos Reis, R.M.; Furtado, C.L.M.; Ferriani, R.A. MIR146A and ADIPOQ genetic variants are associated with birth weight in relation to gestational age: A cohort study. J. Assist. Reprod Genet. 2022, 39, 1873–1886. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, M.; Xu, Q.; Li, J.; Xu, S.; Tang, C. Micro-RNAs in Human Placenta: Tiny Molecules, Immense Power. Molecules 2022, 27, 5943. https://doi.org/10.3390/molecules27185943
Jin M, Xu Q, Li J, Xu S, Tang C. Micro-RNAs in Human Placenta: Tiny Molecules, Immense Power. Molecules. 2022; 27(18):5943. https://doi.org/10.3390/molecules27185943
Chicago/Turabian StyleJin, Meiyuan, Qiang Xu, Jiayong Li, Shouying Xu, and Chao Tang. 2022. "Micro-RNAs in Human Placenta: Tiny Molecules, Immense Power" Molecules 27, no. 18: 5943. https://doi.org/10.3390/molecules27185943
APA StyleJin, M., Xu, Q., Li, J., Xu, S., & Tang, C. (2022). Micro-RNAs in Human Placenta: Tiny Molecules, Immense Power. Molecules, 27(18), 5943. https://doi.org/10.3390/molecules27185943






